Visualising Connections in the Human Brain
Most of us are familiar with pictures from magnetic resonance imaging (MRI) of the human brain; indeed, these black-and-white images have achieved almost iconic status at this stage. From popular television programmes, and regrettably from common experience, the use of these images to detect lesions, such as tumours or the effects of stroke, is well known.
Classic MRI can distinguish grey and white matter based on their different cellular composition but cannot go very far beyond that, because all the white matter has effectively the same contrast. This makes traditional MRI of limited use in examining connectivity between areas of the brain, except at a very gross level (such as whether the corpus callosum exists, for example). However, with a few modifications, MRI can be applied to non-invasively interrogate connectivity in the living human brain, with ever-increasing sensitivity. These new techniques are opening up avenues of investigation that have not just tremendous clinical importance but that also promise to make humans a powerful model organism for the study of axon guidance and related neurodevelopmental processes.
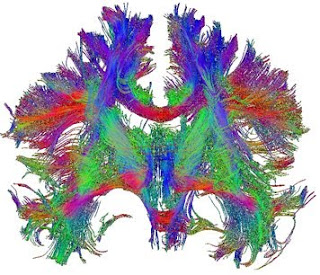
The modifications rely on the fact that the diffusion of water molecules gives off a magnetic resonance signal. Within the brain, the diffusion of water is affected by the local cytoarchitecture; in particular, within axonal bundles the direction of diffusion is constrained by the direction of the axons and their myelin sheaths. By determining the bias in direction of diffusion within a voxel of white matter (the “diffusion tensor”) it is possible to infer the dominant direction of axonal projections in that voxel. Diffusion tensor imaging (DTI) can be applied across the brain, so that, by following the direction of the tensors from voxel to voxel, axonal tracts projecting across large areas of the brain can be derived.
DTI has been extremely powerful, though it does have limitations. Foremost among these is difficulty in distinguishing fibres that cross within a single voxel. More recent refinements, including q-ball imaging and diffusion spectrum imaging (DSI) apply higher-resolution scan acquisition and different statistical approaches to largely resolve this issue. These approaches have yielded dramatic images of fibre tracts within the brain, revealing three-dimensional connectivity patterns across the entire brain that would be impossible to obtain with traditional anatomical tracing or histology, even in post mortem tissue.

It is important to realize, however, that the “fibres” being drawn are really three-dimensional plots of statistical values that may vary depending on method of acquisition, scan parameters, software used, statistical approach as well as subjective thresholding criteria. Comparison of imaging methods and validation using data derived by classical techniques was thus crucial. These kinds of comparisons have now shown that, in the case of DSI at least, the congruence with known or classically-derived tractography is actually very good (Schmahmann et al., 2007; Wedeen et al. , 2008). This is not to say that the approach does not still have limitations – tracking fibres through sharp bends, as they de-fasciculate into small bundles or as they project into grey matter are all still problematic, for example – but these limitations are likely to be overcome by more sophisticated algorithms.
Given the recent pace of improvements in tractography approaches, we can thus expect in the very near future that this technique will allow routine examination of patterns of connectivity in the human brain. These approaches are already being applied to investigate structural connectivity in various disorders with a neurodevelopmental etiology, including schizophrenia, autism, dyslexia and several less well-known conditions, such as prosopagnosia (the inability to recognize faces) and synaesthesia (where sensory stimuli in one modality can cross-activate another).

Tractography can also be used to define a connectivity profile or fingerprint of a particular part of the brain, and thus to automatically parcellate the brain into units of likely functional distinction. This kind of approach is especially useful to delineate functional areas of the neocortex, which are often not obviously anatomically distinct. By defining a connectivity profile of each voxel to all other voxels in the brain, and constructing a matrix of such profiles, it is possible, using a clustering algorithm similar to those applied to microarray data, to cluster voxels with similar connectivity profiles and thus to distinguish regions where the profile suddenly changes, thus delineating the border between two presumptive areas. Exactly this approach has been used by several groups and validated with functional imaging and other anatomical data (Klein et al., 2007; Perrin et al., 2008).
The development of these techniques finally gives us the means to see, literally, differences in how each of our brains is wired, which may affect many aspects of our personality, cognitive abilities and style and other psychological traits that make us who we are.
Wedeen, V., Wang, R., Schmahmann, J., Benner, T., Tseng, W., Dai, G., Pandya, D., Hagmann, P., D'Arceuil, H., & de Crespigny, A. (2008). Diffusion spectrum magnetic resonance imaging (DSI) tractography of crossing fibers NeuroImage, 41 (4), 1267-1277 DOI: 10.1016/j.neuroimage.2008.03.036
Schmahmann, J., Pandya, D., Wang, R., Dai, G., D'Arceuil, H., de Crespigny, A., & Wedeen, V. (2007). Association fibre pathways of the brain: parallel observations from diffusion spectrum imaging and autoradiography Brain, 130 (3), 630-653 DOI: 10.1093/brain/awl359
Perrin, M., Cointepas, Y., Cachia, A., Poupon, C., Thirion, B., Rivière, D., Cathier, P., El Kouby, V., Constantinesco, A., Le Bihan, D., & Mangin, J. (2008). Connectivity-Based Parcellation of the Cortical Mantle Using q-Ball Diffusion Imaging International Journal of Biomedical Imaging, 2008, 1-19 DOI: 10.1155/2008/368406
Klein, J., Behrens, T., Robson, M., Mackay, C., Higham, D., & Johansen-Berg, H. (2007). Connectivity-based parcellation of human cortex using diffusion MRI: Establishing reproducibility, validity and observer independence in BA 44/45 and SMA/pre-SMA NeuroImage, 34 (1), 204-211 DOI: 10.1016/j.neuroimage.2006.08.022
Classic MRI can distinguish grey and white matter based on their different cellular composition but cannot go very far beyond that, because all the white matter has effectively the same contrast. This makes traditional MRI of limited use in examining connectivity between areas of the brain, except at a very gross level (such as whether the corpus callosum exists, for example). However, with a few modifications, MRI can be applied to non-invasively interrogate connectivity in the living human brain, with ever-increasing sensitivity. These new techniques are opening up avenues of investigation that have not just tremendous clinical importance but that also promise to make humans a powerful model organism for the study of axon guidance and related neurodevelopmental processes.

The modifications rely on the fact that the diffusion of water molecules gives off a magnetic resonance signal. Within the brain, the diffusion of water is affected by the local cytoarchitecture; in particular, within axonal bundles the direction of diffusion is constrained by the direction of the axons and their myelin sheaths. By determining the bias in direction of diffusion within a voxel of white matter (the “diffusion tensor”) it is possible to infer the dominant direction of axonal projections in that voxel. Diffusion tensor imaging (DTI) can be applied across the brain, so that, by following the direction of the tensors from voxel to voxel, axonal tracts projecting across large areas of the brain can be derived.
DTI has been extremely powerful, though it does have limitations. Foremost among these is difficulty in distinguishing fibres that cross within a single voxel. More recent refinements, including q-ball imaging and diffusion spectrum imaging (DSI) apply higher-resolution scan acquisition and different statistical approaches to largely resolve this issue. These approaches have yielded dramatic images of fibre tracts within the brain, revealing three-dimensional connectivity patterns across the entire brain that would be impossible to obtain with traditional anatomical tracing or histology, even in post mortem tissue.

It is important to realize, however, that the “fibres” being drawn are really three-dimensional plots of statistical values that may vary depending on method of acquisition, scan parameters, software used, statistical approach as well as subjective thresholding criteria. Comparison of imaging methods and validation using data derived by classical techniques was thus crucial. These kinds of comparisons have now shown that, in the case of DSI at least, the congruence with known or classically-derived tractography is actually very good (Schmahmann et al., 2007; Wedeen et al. , 2008). This is not to say that the approach does not still have limitations – tracking fibres through sharp bends, as they de-fasciculate into small bundles or as they project into grey matter are all still problematic, for example – but these limitations are likely to be overcome by more sophisticated algorithms.
Given the recent pace of improvements in tractography approaches, we can thus expect in the very near future that this technique will allow routine examination of patterns of connectivity in the human brain. These approaches are already being applied to investigate structural connectivity in various disorders with a neurodevelopmental etiology, including schizophrenia, autism, dyslexia and several less well-known conditions, such as prosopagnosia (the inability to recognize faces) and synaesthesia (where sensory stimuli in one modality can cross-activate another).

Tractography can also be used to define a connectivity profile or fingerprint of a particular part of the brain, and thus to automatically parcellate the brain into units of likely functional distinction. This kind of approach is especially useful to delineate functional areas of the neocortex, which are often not obviously anatomically distinct. By defining a connectivity profile of each voxel to all other voxels in the brain, and constructing a matrix of such profiles, it is possible, using a clustering algorithm similar to those applied to microarray data, to cluster voxels with similar connectivity profiles and thus to distinguish regions where the profile suddenly changes, thus delineating the border between two presumptive areas. Exactly this approach has been used by several groups and validated with functional imaging and other anatomical data (Klein et al., 2007; Perrin et al., 2008).
The development of these techniques finally gives us the means to see, literally, differences in how each of our brains is wired, which may affect many aspects of our personality, cognitive abilities and style and other psychological traits that make us who we are.
Wedeen, V., Wang, R., Schmahmann, J., Benner, T., Tseng, W., Dai, G., Pandya, D., Hagmann, P., D'Arceuil, H., & de Crespigny, A. (2008). Diffusion spectrum magnetic resonance imaging (DSI) tractography of crossing fibers NeuroImage, 41 (4), 1267-1277 DOI: 10.1016/j.neuroimage.2008.03.036
Schmahmann, J., Pandya, D., Wang, R., Dai, G., D'Arceuil, H., de Crespigny, A., & Wedeen, V. (2007). Association fibre pathways of the brain: parallel observations from diffusion spectrum imaging and autoradiography Brain, 130 (3), 630-653 DOI: 10.1093/brain/awl359
Perrin, M., Cointepas, Y., Cachia, A., Poupon, C., Thirion, B., Rivière, D., Cathier, P., El Kouby, V., Constantinesco, A., Le Bihan, D., & Mangin, J. (2008). Connectivity-Based Parcellation of the Cortical Mantle Using q-Ball Diffusion Imaging International Journal of Biomedical Imaging, 2008, 1-19 DOI: 10.1155/2008/368406
Klein, J., Behrens, T., Robson, M., Mackay, C., Higham, D., & Johansen-Berg, H. (2007). Connectivity-based parcellation of human cortex using diffusion MRI: Establishing reproducibility, validity and observer independence in BA 44/45 and SMA/pre-SMA NeuroImage, 34 (1), 204-211 DOI: 10.1016/j.neuroimage.2006.08.022



Nice post - pictures of the human brain ..Keep Posting
ReplyDeleteRon
pictures of the human brain
Thanks for the post. I was always interesting in the question of visualizing connection in our brain. Today I know the answer and I must say that you know it, too))) Owing to custom writing services I had a very good article on this topic!
ReplyDeleteThis is fantastic, there is so much of our brain that we just dont know or understand, we know more about oceans and space than our brain capabilities.
ReplyDeletexl pharmacy
I am going to keep it in your mind, appreciate it for giving the details keep upgrading, looking forward for additional posts.
ReplyDeleteShuvo,
Clipping Path
Your blog it gave me a lot of fun, I learned a lot from your blog you are looking forward to many more articles.
ReplyDeleteShuvo,
Clipping Path
I think more updates and will be returning. I have filtered for qualified edifying substance of this calibre all through the past various hours. www.hastings-online.co.uk |
ReplyDeletewww.steppescaribbean.co.uk |
www.hawnbyhotel.co.uk |
www.partridgeplace.co.uk |
www.newelfin.co.uk |
www.subito2.co.uk |
www.kingsofbassline.co.uk |
www.beckyandneil.co.uk |
www.antichew.co.uk |
www.oldtimegrates.co.uk |